Mutagenesis of
Trp54 and Trp203 Residues on Fibrobacter
Succinogenes 1,3-1,4-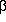 -D-Glucanase
Significantly Affects Catalytic Activities of the Enzyme -D-Glucanase
Significantly Affects Catalytic Activities of the Enzyme
Hsueh-Ling Cheng,
Li-Chu Tsai,
Su-Shiang Lin,
Hanna S. Yuan,
Ning-Sun Yang,
Shu-Hua Lee, and
Lie-Fen Shyur
Institute of BioAgricultural
Sciences, Academia Sinica, Taiwan, R.O.C., Department of Nursing, Mei Ho
Institute of Technology, Taiwan, R.O.C., and Institute of Molecular
Biology, Academia Sinica, Taiwan, R.O.C.
Abstract:
The possible structural and catalytic
functions of the nine tryptophan amino acid residues, including Trp54,
Trp105, Trp112, Trp141, Trp148,
Trp165, Trp186, Trp198, and Trp203
in Fibrobacter succinogenes 1,3-1,4-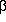 -D-glucanase
(Fs -D-glucanase
(Fs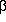 -glucanase), were
characterized using site-directed mutagenesis, initial rate kinetics,
fluorescence spectrometry, and structural modeling analysis. Kinetic
studies showed that a 5-7-fold increase in Km value
for lichenan was observed for W141F, W141H, and W203R mutant Fs -glucanase), were
characterized using site-directed mutagenesis, initial rate kinetics,
fluorescence spectrometry, and structural modeling analysis. Kinetic
studies showed that a 5-7-fold increase in Km value
for lichenan was observed for W141F, W141H, and W203R mutant Fs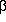 -glucanases,
and approximately 72-, 56-, 30-, 29.5-, 4.9-, and 4.3-fold decreases in kcat
relative to that for the wild-type enzyme were observed for the W54F,
W54Y, W141H, W203R, W141F, and W148F mutants, respectively. In contrast,
W186F and W203F, unlike the other 12 mutants, exhibited a 1.4- and
4.2-fold increase in kcat, respectively. W165F and
W203R were the only two mutants that exhibited a 4-7-fold higher
activity relative to the wild-type enzyme after they were incubated at
pH 3.0 for 1 h. Fluorescence spectrometry indicated that all of the
mutations on the nine tryptophan amino acid residues retained a folding
similar to that of the wild-type enzyme. Structural modeling and kinetic
studies suggest that Trp54, Trp141, Trp148,
and Trp203 play important roles in maintaining structural
integrity in the substrate-binding cleft and the catalytic efficiency of
the enzyme. -glucanases,
and approximately 72-, 56-, 30-, 29.5-, 4.9-, and 4.3-fold decreases in kcat
relative to that for the wild-type enzyme were observed for the W54F,
W54Y, W141H, W203R, W141F, and W148F mutants, respectively. In contrast,
W186F and W203F, unlike the other 12 mutants, exhibited a 1.4- and
4.2-fold increase in kcat, respectively. W165F and
W203R were the only two mutants that exhibited a 4-7-fold higher
activity relative to the wild-type enzyme after they were incubated at
pH 3.0 for 1 h. Fluorescence spectrometry indicated that all of the
mutations on the nine tryptophan amino acid residues retained a folding
similar to that of the wild-type enzyme. Structural modeling and kinetic
studies suggest that Trp54, Trp141, Trp148,
and Trp203 play important roles in maintaining structural
integrity in the substrate-binding cleft and the catalytic efficiency of
the enzyme.
|