Redesign of High-Affinity Nonspecific Nucleases with Altered Sequence Preference
Yi-Ting Wang,†,‡,§ Jon D. Wright,| Lyudmila G. Doudeva,† Hua-Ci Jhang,†
Carmay Lim,*,|,‡,§ and Hanna S. Yuan*,†,‡,#
| Institute of Molecular Biology, Institute of Biomedical Sciences, and Institute of Chemical Biology and Molecular Biophysics, Taiwan International Graduate Program, Academia Sinica, Taipei, Taiwan, R.O.C, Institute of Bioinformatics and Structural Biology and Department of Chemistry, National Tsing Hua UniVersity, HsinChu, Taiwan, R.O.C., and Institute of Biochemistry and Molecular Biology, National Taiwan UniVersity, Taipei, Taiwan, R.O.C Received August 24, 2009; E-mail: hanna@sinica.edu.tw; carmay@gate.sinica.edu.tw Abstract: It is of crucial importance to elucidate the underlying principles that govern the binding affinity and selectivity between proteins and DNA. Here we use the nuclease domain of Colicin E7 (nColE7) as a model system to generate redesigned nucleases with improved DNA-binding affinities. ColE7 is a bacterial toxin, bearing a nonspecific endonuclease domain with a preference for hydrolyzing DNA phosphodiester bonds at the 3′O-side after thymine and adenine; i.e., it prefers Thy and Ade at the -1 site. Using systematic computational screening, six nColE7 mutants were predicted to bind DNA with high affinity. Five of the redesigned single-point mutants were constructed and purified, and four mutants had a 3- to 5-fold higher DNA binding affinity than wild-type nColE7 as measured by fluorescence kinetic assays. Moreover, three of the designed mutants, D493N, D493Q, and D493R, digested DNA with an increased preference for guanine at +3 sites compared to the wild-type enzyme, as shown by DNA footprint assays. X-ray structure determination of the ColE7 mutant D493Q-DNA complex in conjunction with structural and free energy decomposition analyses provides a physical basis for the improved protein-DNA interactions: Replacing D493 at the protein-DNA interface with an amino acid residue that can maintain the native hydrogen bonds removes the unfavorable electrostatic repulsion between the negatively charged carboxylate and DNA phosphate groups. These results show that computational screening combined with biochemical, structural, and free energy analyses provide a useful means for generating redesigned nucleases with a higher DNA-binding affinity and altered sequence preferences in DNA cleavage. | 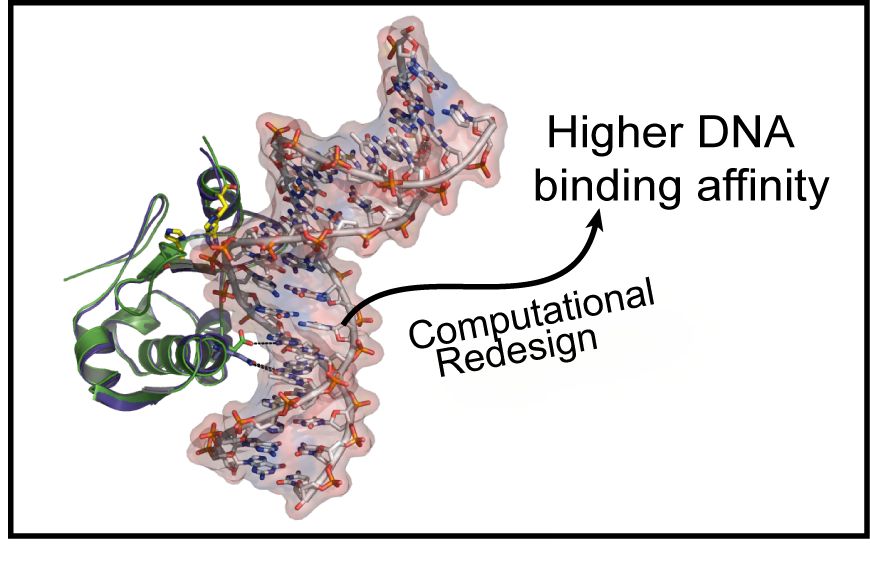 |