| Structural insights into TDP-43 in nucleic-acid binding and domain interactions |
| Pan-Hsien Kuo1,2, Lyudmila G. Doudeva2, Yi-Ting Wang1,2,3, Che-Kun James Shen2 and Hanna S. Yuan2,3,4,* 1Institute of Bioinformatics and Structural Biology, National Tsing Hua University, 2Institute of Molecular Biology, 3Taiwan International Graduate Program, Chemical Biology and Molecular Biophysics, Academia Sinica and 4Graduate Institute of Biochemistry and Molecular Biology, National Taiwan University, Taipei, Taiwan, ROC Received October 14, 2008; Revised November 27, 2008; Accepted January 7, 2009 TDP-43
is a pathogenic protein: its normal function in binding to UG-rich RNA
is related to cystic fibrosis, and inclusion of its C-terminal
fragments in brain cells is directly linked to frontotemporal lobar
degeneration (FTLD) and amyotrophic lateral sclerosis (ALS). Here we
report the 1.65A ° crystal structure of the C-terminal RRM2 domain of
TDP-43 in complex
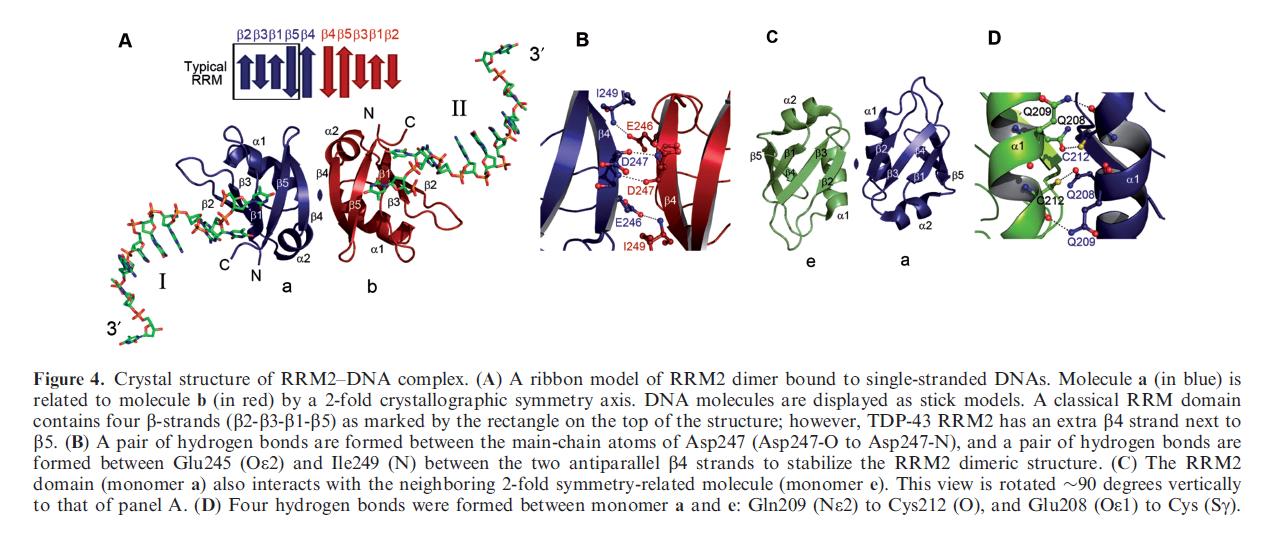
with a single-stranded DNA. We show that TDP-43 is a dimeric protein with two RRM domains, both involved in DNA and RNA binding. The crystal structure reveals the basis of TDP-43’s TG/UG preference in nucleic acids binding. It also reveals that RRM2 domain has an atypical RRM-fold with an additional b-strand involved in making protein– protein interactions. This self association of RRM2 domains produced thermal-stable RRM2 assemblies with a melting point greater than 858C as monitored by circular dichroism at physiological conditions. These studies thus characterize the recognition between TDP-43 and nucleic acids and the mode of RRM2 self association, and provide molecular models for understanding the role of TDP-43 in cystic fibrosis and the neurodegenerative diseases related to TDP-43 proteinopathy. |