Pan-Hsien Kuo1, Chien-Hao Chiang1,2, Yi-Ting Wang1, Lyudmila G. Doudeva1 and Hanna S. Yuan1,3,*
1Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan, 2Institute of Bioinformatics and Structural
Biology, National Tsing Hua University, Hsin Chu, Taiwan and 3Graduate Institute of Biochemistry and
Molecular Biology, National Taiwan University, Taipei 10048, Taiwan
Received July 2, 2013; Revised November 22, 2013; Accepted December 23, 2013
ABSTRACT
TDP-43 is an important pathological protein that aggregates in the diseased neuronal cells and is linked to various neurodegenerative disorders. In normal cells, TDP-43 is primarily an RNA-binding protein;
however, how the dimeric TDP-43 binds RNA via its two RNA recognition motifs, RRM1 and RRM2, is not clear. Here we report the crystal structure of human TDP-43 RRM1 in complex with a singlestranded DNA showing that RRM1 binds the nucleic acid extensively not only by the conserved b-sheet residues but also by the loop residues.Mutational and biochemical assays further reveal that both RRMs in TDP-43 dimers participate in binding of UG-rich RNA or TG-rich DNA with RRM1 playing a dominant role and RRM2 playing a supporting role. Moreover, RRM1 of the amyotrophic lateral sclerosis-linked mutant D169G binds DNA as efficiently as the wild type; nevertheless, it is more resistant to thermal denaturation, suggesting that the resistance to degradation is likely linked to TDP-43 proteinopathies. Taken together all the data, we suggest a model showing that the two RRMs in each protomer of TDP-43 homodimer work together in RNA binding and thus the dimeric TDP-43 recognizes long clusters of UG-rich RNA to achieve high affinity and specificity.
TDP-43 is an important pathological protein that aggregates in the diseased neuronal cells and is linked to various neurodegenerative disorders. In normal cells, TDP-43 is primarily an RNA-binding protein;
however, how the dimeric TDP-43 binds RNA via its two RNA recognition motifs, RRM1 and RRM2, is not clear. Here we report the crystal structure of human TDP-43 RRM1 in complex with a singlestranded DNA showing that RRM1 binds the nucleic acid extensively not only by the conserved b-sheet residues but also by the loop residues.Mutational and biochemical assays further reveal that both RRMs in TDP-43 dimers participate in binding of UG-rich RNA or TG-rich DNA with RRM1 playing a dominant role and RRM2 playing a supporting role. Moreover, RRM1 of the amyotrophic lateral sclerosis-linked mutant D169G binds DNA as efficiently as the wild type; nevertheless, it is more resistant to thermal denaturation, suggesting that the resistance to degradation is likely linked to TDP-43 proteinopathies. Taken together all the data, we suggest a model showing that the two RRMs in each protomer of TDP-43 homodimer work together in RNA binding and thus the dimeric TDP-43 recognizes long clusters of UG-rich RNA to achieve high affinity and specificity.
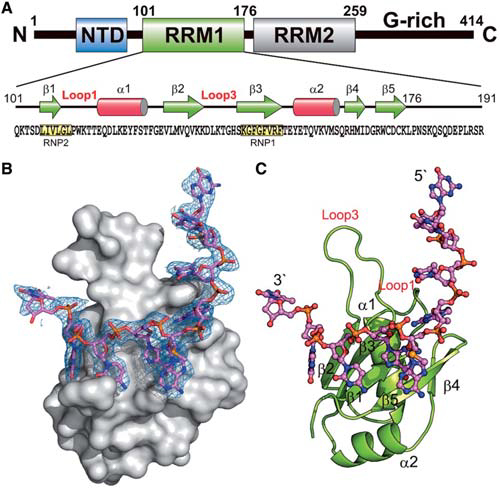
Overall crystal structure of hRRM1-DNA complex