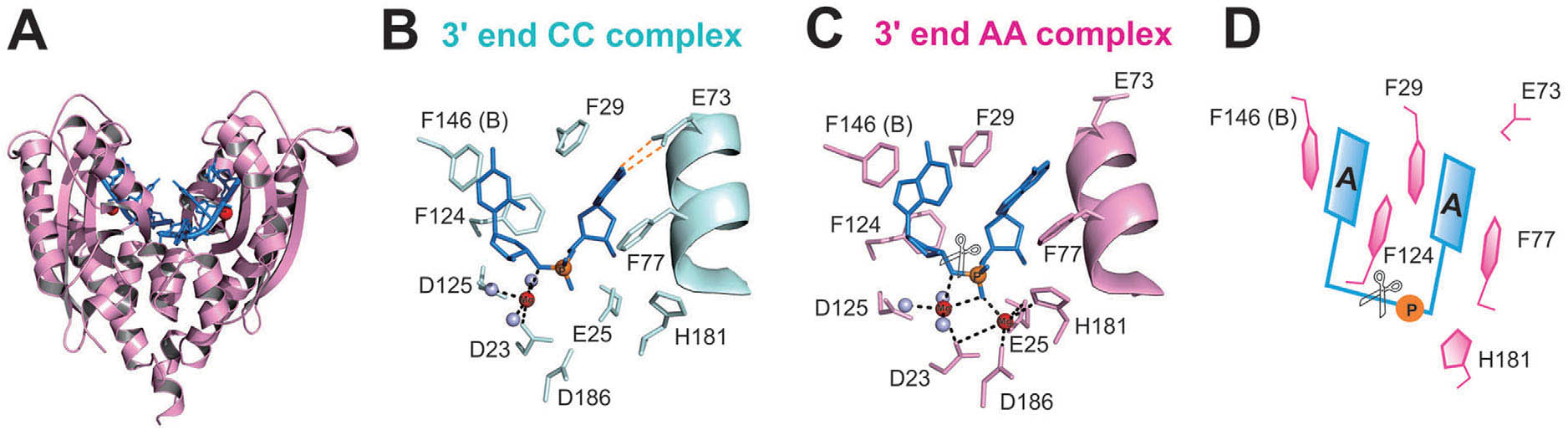
Aromatic residues in RNase T stack with nucleobases to guide the sequence-specific recognition and cleavage of nucleic acids
Yulander Duh,1,2 Yu-Yuan Hsiao,3 Chia-Lung Li,1 Jason C. Huang,2 andHanna S. Yuan1,4*
Abstract: RNase T is a classical member
of the DEDDh family of exonucleases with a unique sequence preference
in that its 30-to-50 exonuclease activity is blocked by a 30-terminal
dinucleotide CC in digesting both single-stranded RNA and DNA. Our
previous crystal structure analysis of RNase T-DNA complexes show that
four phenylalanine residues, F29, F77, F124, and F146, stack with the
two 30-terminal nucleobases. To elucidate if the p–p stacking
interactions between aromatic residues and nucleobases play a critical
role in sequence-specific protein–nucleic acid recognition, here we
mutated two to four of the phenylalanine residues in RNase T to
tryptophan (W mutants) and tyrosine (Y mutants). The Escherichia coli
strains expressing either the W mutants or the Y mutants had slow
growth phenotypes, suggesting that all of these mutants could not fully
substitute the function of the wild-type RNase T in vivo. DNA digestion assays revealed W mutants shared similar sequence specificity with wild-type RNase T. However, the Y mutants exhibited altered sequence-dependent activity, digesting ssDNA with both 30-end CC and GG sequences. Moreover, the W and Y mutants had reduced DNA-binding activity and lower thermal stability as compared to wild-type RNase T. Taken together, our results suggest that the four phenylalanine residues in RNase T not only play critical roles in sequence-specific recognition, but also in overall protein stability. Our results provide the first evidence showing that the p2p stacking interactions between nucleobases and protein aromatic residues may guide the sequence-specific activity for DNA and RNA enzymes.
substitute the function of the wild-type RNase T in vivo. DNA digestion assays revealed W mutants shared similar sequence specificity with wild-type RNase T. However, the Y mutants exhibited altered sequence-dependent activity, digesting ssDNA with both 30-end CC and GG sequences. Moreover, the W and Y mutants had reduced DNA-binding activity and lower thermal stability as compared to wild-type RNase T. Taken together, our results suggest that the four phenylalanine residues in RNase T not only play critical roles in sequence-specific recognition, but also in overall protein stability. Our results provide the first evidence showing that the p2p stacking interactions between nucleobases and protein aromatic residues may guide the sequence-specific activity for DNA and RNA enzymes.
Crystal
structures
of RRM1/DNA and RRM1-D169G/DNA complexes.