Research
Interest
Structural
and functional studies of DNA/RNA degradation nucleases in protection or
killing of cells
Activation of
nucleases may result in nucleic acid degradation and lead to contradictory consequences-
either protection or killing of cells.
A
variety of non-specific nucleases have been
discovered in
prokaryotes and eukaryotes that are
triggered during cell defense or programmed cell death. We are interested in
understanding and comparing the regulation strategies, nucleic acid recognition
methods and catalytic mechanisms of these nucleases using a combination of
structural, biophysical and biochemical approaches.
Two types of
sugar non-specific nucleases have been identified in bacteria to take part in
host defense. The first, a family
of periplasmic nucleases, including Vvn from Vibrio vulnificus, protect the cell by preventing the uptake
of foreign DNA molecules. The Escherichia coli released nuclease-type bacteriacins
represent another class of non-specific endonucleases
which digest nucleic acids randomly in target foreign cells to induce cell
death, thereby improving host cell survival advantage during stress. We have resolved the
crystal structures of Vvn and ColE
We will continue our studies on Vvn and ColE7
and also extend our investigation to the eukaryotic non-specific nucleases that
are involved in RNA degradation and programmed cell death. Functional and structural insight into
this group of non-specific nucleases is valuable for developing strategies to
promote or suppress cell survival machinery.
|
|
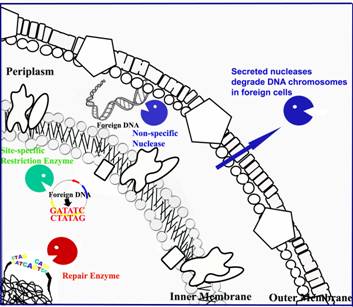
Different
types of nucleases constitute a secure network for bacterial host defense
against foreign nucleic acids.
Restriction enzymes cleave foreign unmethylated
DNA in a site-specific manner. Periplasmic nucleases degrade foreign nucleic acids
non-specifically in periplasmic space. Cells also release nuclease-type
toxins to degrade nucleic acids in foreign cells to increase host survival
advantage. |
|
|
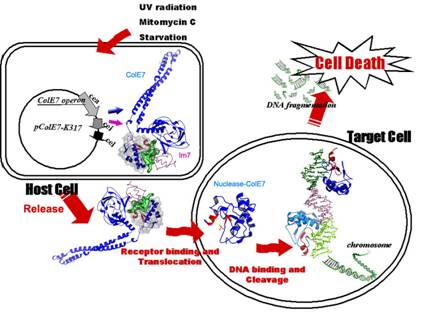
The nuclease-type toxin ColE7 is expressed
in host cells and cleaves nucleic acid molecules in foreign cells. In the host cell, ColE7 and its
inhibitor Im7 are co-expressed and secreted as a hetero-dimeric
complex. ColE7 contains three
functional domains, receptor binding, membrane translocation and nuclease
domains. After ColE7 binds to the
receptor on foreign cells, ColE7 is cleaved during translocation. Only the nuclease domain of ColE7
reaches the cytoplasm of foreign cells for nucleic acid degradation. |
|
|
The crystal structures of two bacterial
sugar-nonspecific nucleases, Vvn and the nuclease
domain of ColE7. The active sites
(displaying in red) of the two enzymes contain a common bba-metal
topology (His-metal finger). Upon
DNA binding, the bba-metal fold binds at the minor
groove and induces DNA bending slightly away from the enzyme. |