Research Interest
Structural
studies of nucleic acids-binding proteins in translation regulation and nucleic
acid degradation
We
are interested in a number of proteins involved
in translation regulation and DNA/RNA degradation. The overall goal is to discover
structure-based mechanisms of these proteins in nucleic acids recognition and
degradation. We use
a major tool of X-ray crystallography in combination with mutagenesis,
biochemical and biophysical approaches. Several projects of interest are listed
below.
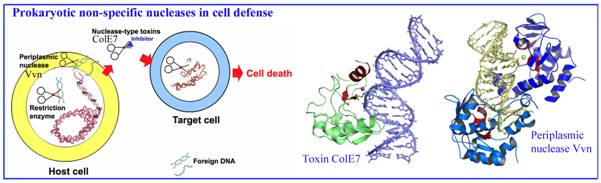
1. Bacterial nucleases in cell defense
We have been working on two types of sugar
non-specific nucleases in bacteria, including a periplasmic nuclease Vvn and a secreted toxin ColE7, both of which digest
foreign nucleic acids for cell defense. Based on our structural and biochemical analysis on Vvn
and ColE7, we have provided a solid foundation to explain how these nucleases
are inhibited and activated, how they recognize DNA without sequence
specificity and how they digest DNA to protect bacterial cells at atomic level.
References:
Li, C., Ho, L.-I., Chang, Z.-F., Tsai, L.-C., Yang,
W.-Z. and Yuan*, H. S. (2003) DNA binding and cleavage by the periplasmic
nuclease Vvn: A novel structure with a known active site. EMBO J. 22, 4014-4025.
Hsia, K.-C., Chak, K.-F., Liang, P.-H., Cheng, Y.-S.,
Ku, W.-Y. and Yuan*, H. S. (2004) DNA binding
and degradation by the H-N-H protein ColE7. Structure
12, 205-214.
Hsia, K.-C., Li, C.-L. and Yuan*, H. S. (2005) Structural and functional insight
into the sugar-nonspecific nucleases in host defense, Curr. Opin. Struct. Biol. 15, 126-134.
Shi, Z., Chak, K.-F. and Yuan*, H. S. (2005) Identification of an essential
cleavage site in ColE7 required for import and killing cells, J. Biol. Chem. 26, 24663-24668.
Cheng, Y.-S., Shi, Z., Doudeva, L. G., Yang, W.-Z., Chak, K.-F. and Yuan*, H.
S. (2006) High-resolution crystal structure of a
truncated ColE7 translocation domain: Implications for colicin transport across
membranes. J. Mo. Biol., 356, 22-31.
Wang,
Y.-T., Yang, W.-J., Li, C.-L., Doudeva,
L. G. and Yuan*, H. S. (2007) Structural basis for sequence-dependent cleavage by nonspecific endonucleases. Nucleic Acid Res. 35, 584-594.
2. Tudor-SN in miRNA degradation and mRNA translation regulation
Tudor-SN is a multifunctional protein, playing
a role in transcription regulation, RNA editing, interference and splicing.
Recent studies show that Tudor-SN is a miRNase specific for inosine-containing
microRNA precursors, and it also regulates gene expression by binding to mRNA
at
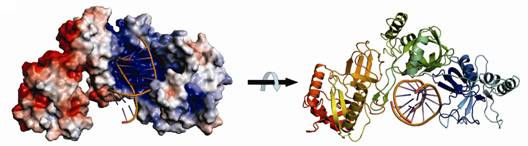
Structural model of a
64-kD Tudor-SN bound to double-stranded RNA.
Our biochemical and structural data suggest that tandem repeats of SN domains in
Tudor-SN work together to capture RNA substrates.
References:
Li,
C.-L., Yang, W.-Z., Chen, Y.-P. and Yuan*, H. S. (2008) Structural and
functional insights into human Tudor-SN, a key component linking RNA
interference and editing. Nucleic Acid Res. 36, 3579-3589.
3. PNPase in mRNA degradation
PNPase (polynucleotide phosphorylase) is
an important enzyme responsible for mRNA turnover from
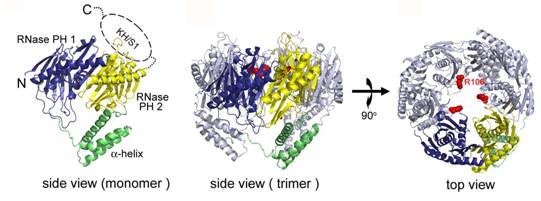
Reference:
Shi, Z., Yang, W.-Z., Lin-Chao, S., Chak, K.-F. and Yuan*, H. S. Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive
RNA degradation. RNA (in press).
4.
Apoptotic nucleases in DNA degradation.
Apoptotic nucleases are activated for chromosomal DNA fragmentation during apoptosis. Inactivation
of these apoptotic nucleases produces undigested DNA and is related to a number
of autoimmune disorders. We analyze the biochemical properties and crystal
structures of a number of apoptotic nucleases to address the function of these
nucleases in normal versus apoptotic cells. Recently, we determined the crystal
structure of a C. elegans
cell-death-related nuclease 4 (CRN-4). The biochemical,
structural, and functional assays consistently suggest that the C-terminal
novel-fold Zn-domain of CRN-4 is involved in DNA binding and the N-terminal
nuclease domain is responsible for DNA degradation. This study therefore
provides new insights into the DEDDh family of nucleases in chromosomal DNA
fragmentation in apoptosis.
We also analyze the biochemical and structural
features of several apoptotic proteins and nucleases that interact with CRN-4 to
form a degradeosome in apoptosis, including CPS-6 (human Endo G homologue),
WAH-1 (AIF), CRN-5 (Rrp46) and Cyp-13. The long-term goal of this research is
to decipher the working mechanism of the degradeosome in DNA fragmentation
during apoptosis.
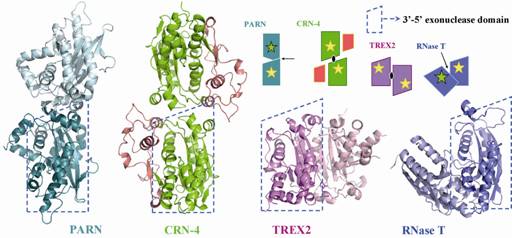
Crystal structure of CRN-4 and Comparison of the different domain arrangement in dimeic DEDDh family
proteins. CRN-4 dimerizes in a different mode as compared to PARN, TREX2 and
RNase T.
Reference:
Hsiao, Y. Y., Nakagawa, A., Shi,
Z., Mitani, S., Xue, D. and
Yuan*, H. S. Crystal structure of CRN-4: implications for domain
function in apoptotic DNA degradation. (under revision).