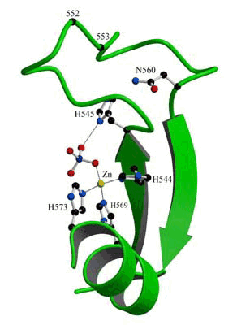
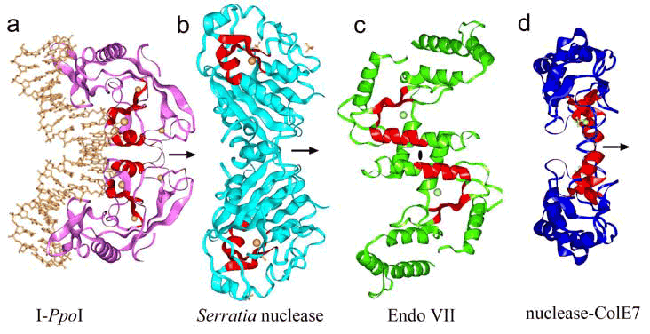
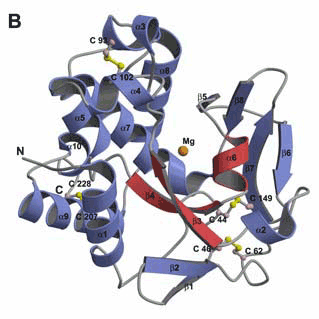
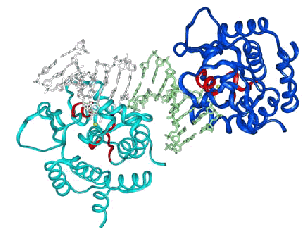
HNH consensus E HH P GG NL H H
Bacteriocins Colicin E7 SGKRTSFELHHEKPISQNGGVYDMDNISVVTPKRHIDIHRGK-576
Colicin E2 VGGRERFELHHDKPISQDGGVYDMNNIRVTTPKRHIDIHRGK-581
Colicin E8 VGGRRSFELHHDKPISQDGGVYDMDNLRITTPKRHIDIHRGQ
Colicin E9 VGGRVKYELHHDKPISQGGEVYDMDNIRVTTPKRHIDIHRGK-582
Pyocin S1 AGGRIKIEIHHKVRVADGGGVYNMGNLVAVTPKRHIEIHKGG-617
Pyocin S2 AGGRIKIEIHHKVRIADGGGVYNMGNLVAVTPKRHIEIHKGG-770
Group I homing I-HmuI EGYEEGLVVDHKD...GNKDNNLSTNLRWVTQKINVENQMSR-77
endonuclease I-HmuII GGYEESLVVDHID...RNRHNNHFSNLRWVSRKENSSNISAD-104
I-HmuIII EGYGEDLVVDHID...QDRDNNHCSNLRWVSRKENSNNISAD-103
yosQ YDIPKGMFVNHID...GNKLNNHVRNLEIVTPKENTLHAMKI-102
I-TevIII DSDGRTDEIHHKD...GNRENNDLDNLMCLSIQEHYDIHLAQ-55
ORF253 VT-DKNKYIDHIN...GNPLDNRRNNLRVVSHQENMMNKKTY-192
Group II homing Avi CPMC9 WNIHHIIKRHMGGGDEL.DNLVLLHPNCHRQLH
endonuclease Cpc1 CSHC9 IEIDHIIP.KSQGGKDVYDNLQALHRHCHDVKTATD-568
Cpc2 CSEC9 MEVHHIDQNR..GNNKL.SNLTLVHRHCHDIIH
PetD INSIP.YELHHILP.KRFGGKDTPNNMVLLCKSPCHQLVSSSI-574
Restriction or McrA CENC14LEVHHVIP.LSSGGADTTDNCVALCPNCHRELHYSK-259
Repair enzyme S.g. mtMSH ICGAPADAVHHIKP6LCNRKLNRRSNLVPVCSSCHLDIHRNK-953
|